TF symbolic engine#
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/jdariasl/OTBD/main/content/init.py
import init; init.init(force_download=False)
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
%load_ext tensorboard
from sklearn.datasets import *
from local.lib.utils import make_graph
from IPython.display import Image
tf.__version__
2024-12-10 17:51:54.117859: I tensorflow/core/platform/cpu_feature_guard.cc:182] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
'2.14.0'
TF is a symbolic computing + optimization library for machine learning problems#
ML expressions involve:
variables representing data as n-dimensional objects
variables representing parameters as n-dimensional objects
mostly matrix operations (multiplications, convolutions, etc.)
some non linear operations (activation functions)
Recall that in sympy we FIRST define expressions (a computational graph) and THEN we evaluate them feed concrete values.
Tensorflow INTEGRATES both aspects so that building computational graphs LOOKS LIKE writing regular Python code as must as possible.
a
tf.Variablerepresents a symbolic variable, that contains a value
See:
https://www.tensorflow.org/guide/keras/custom_layers_and_models
https://www.tensorflow.org/guide/keras/customizing_what_happens_in_fit
x = tf.Variable(initial_value=[7], name="x", dtype=tf.float32)
y = tf.Variable(initial_value=[9], name="y", dtype=tf.float32)
f = x**2+y**3
f
<tf.Tensor: shape=(1,), dtype=float32, numpy=array([778.], dtype=float32)>
f is SYMBOLIC EXPRESSION (a Tensor in TF terms) that also contains a value attached to it.
for which TF can obtain gradients automatically. This might seem a rather akward way of obtaining the gradient (with GradientTape). The goal is that you write code as in Python and TF takes care of building the computational graph with it.
with tf.GradientTape(persistent=True) as t:
f = x**2 + y**3
print (t.gradient(f, x), t.gradient(f, y))
tf.Tensor([14.], shape=(1,), dtype=float32) tf.Tensor([243.], shape=(1,), dtype=float32)
print (t.gradient(f, [x,y]))
[<tf.Tensor: shape=(1,), dtype=float32, numpy=array([14.], dtype=float32)>, <tf.Tensor: shape=(1,), dtype=float32, numpy=array([243.], dtype=float32)>]
usually expressions are built within functions decorated with @tf.function for performance
@tf.function
def myf(x,y):
return x**2 + y**3
with tf.GradientTape(persistent=True) as t:
f = myf(x,y)
print (t.gradient(f, x), t.gradient(f, y))
tf.Tensor([14.], shape=(1,), dtype=float32) tf.Tensor([243.], shape=(1,), dtype=float32)
!rm -rf logs
make_graph(myf, x, y, logdir="logs")
<tensorflow.python.eager.polymorphic_function.polymorphic_function.Function object at 0x7fce22dc06d0>
%tensorboard --logdir logs
Tensors#
in Tensorflow the notion of a Tensor is just a symbolic multidimensional array.
Observe how Tensorflow naturally deals with multidimensional symbolic variables (Tensors)
n = 3
X = tf.Variable(initial_value=[[2, 6], [3, 1], [4, 5]], name="X", dtype=tf.float32)
w = tf.Variable(initial_value=[[-2],[1]], name="w", dtype=tf.float32)
y = tf.Variable(initial_value=[[8],[2],[3]], name="y", dtype=tf.float32)
with tf.GradientTape(persistent=True) as t:
f = tf.reduce_mean((tf.matmul(X,w)-y)**2)
g = t.gradient(f, w)
g
<tf.Tensor: shape=(2, 1), dtype=float32, numpy=
array([[-38. ],
[-48.666668]], dtype=float32)>
But a tf.Tensor is always a symbolic variable. In order to reconcile symbolic and execution worlds, Tensorflow attaches a value to each symbolic variable, and carries it forward when making derivations.
X,yandware Tensors that we define with a specific valuegis a Tensor derived fromX,yandwthat have ALSO been evaluated with the corresponding values.
g.numpy()
array([[-38. ],
[-48.666668]], dtype=float32)
Note that you can also estimate derivatives with respect to data:
t.gradient(f, X)
<tf.Tensor: shape=(3, 2), dtype=float32, numpy=
array([[ 8. , -4. ],
[ 9.333334, -4.666667],
[ 8. , -4. ]], dtype=float32)>
Implementing linear regresion in TF#
d = pd.read_csv("local/datasets/trilotropicos.csv")
y = d.densidad_escamas.values.astype(np.float32)
X = np.r_[[d.longitud.values]].T.astype(np.float32)
print(X.shape, y.shape)
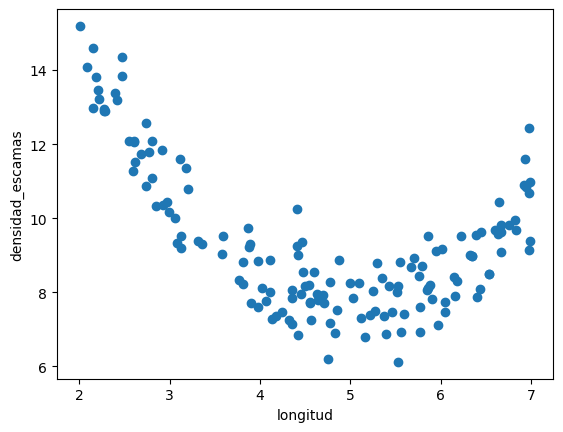
plt.scatter(d.longitud, d.densidad_escamas)
plt.xlabel(d.columns[0])
plt.ylabel(d.columns[1])
(150, 1) (150,)
Text(0, 0.5, 'densidad_escamas')
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X,y)
lr.coef_, lr.intercept_
(array([-0.71805906], dtype=float32), 12.689999)
Version 1: raw low level with gradient descent#
beware of typing.
tensorflowis very sensitive to numeric data types (tf.float32,tf.float64, etc.) Default types innumpyandtensorflowmight not always be the same
from tqdm import tqdm
epochs = 4000
learning_rate = 0.01
# symbolic variables
w = tf.Variable(np.random.normal(size=(X.shape[-1], 1), scale=.6), dtype=tf.float32)
b = tf.Variable(np.random.normal(size=(1,), scale=.6), dtype=tf.float32)
h = []
#optimization loop
for epoch in tqdm(range(epochs)):
with tf.GradientTape() as t:
preds = tf.matmul(X,w)+b
loss = tf.reduce_mean( (preds-y.reshape(-1,1))**2)
gw, gb = t.gradient(loss, [w, b])
w.assign_sub(learning_rate * gw)
b.assign_sub(learning_rate * gb)
h.append([gw.numpy()[0][0], gb.numpy()[0], w.numpy()[0][0], b.numpy()[0], loss.numpy()])
h = np.r_[h]
print (b.numpy(), w.numpy())
100%|████████████████████████████████████| 4000/4000 [00:07<00:00, 500.94it/s]
[12.678262] [[-0.7157532]]
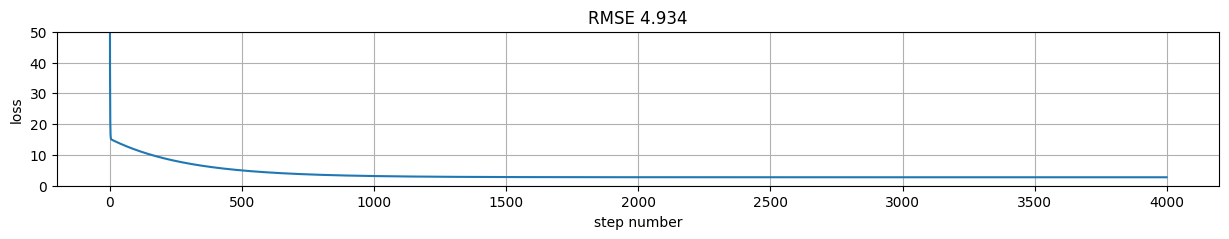
rmse = tf.reduce_mean((tf.matmul(X,w)+b-y)**2).numpy()
plt.figure(figsize=(15,2));
plt.plot(h[:,-1]); plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse);
plt.ylim(0,50)
(0.0, 50.0)
Version 2: using tf.function to speed up#
epochs = 4000
# initialize weights
w.assign(np.random.normal(size=(X.shape[-1],1)).astype(np.float32)*.6)
b.assign(np.random.normal(size=(1,)).astype(np.float32))
@tf.function
def get_gradient(w, b, X, y):
with tf.GradientTape() as t:
preds = tf.matmul(X,w)+b
loss = tf.reduce_mean( (preds-y)**2)
gw, gb = t.gradient(loss, [w, b])
return gw, gb, loss
#optimization loop
h = []
for epoch in tqdm(range(epochs)):
gw, gb, loss = get_gradient(w, b, X, y.reshape(-1,1))
w.assign_sub(learning_rate * gw)
b.assign_sub(learning_rate * gb)
h.append([gw.numpy()[0][0], gb.numpy()[0], w.numpy()[0][0], b.numpy()[0], loss.numpy()])
h = np.r_[h]
print (b.numpy(), w.numpy())
100%|████████████████████████████████████| 4000/4000 [00:04<00:00, 816.77it/s]
[12.677817] [[-0.7156658]]
predictions = tf.matmul(X,w)+b
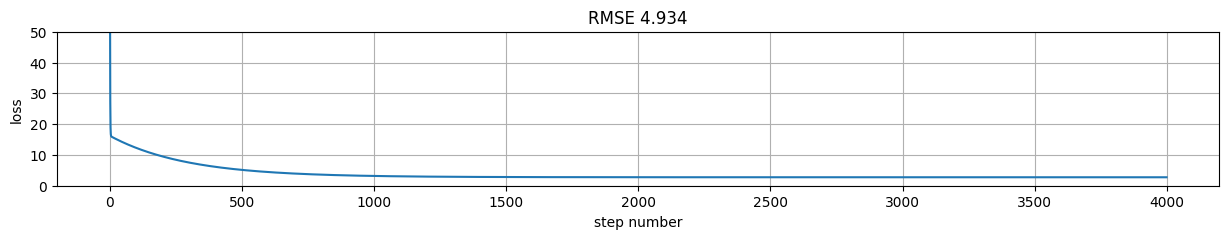
rmse = tf.reduce_mean((predictions-y)**2).numpy()
plt.figure(figsize=(15,2));
plt.plot(h[:,-1]); plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse);
plt.ylim(0,50)
(0.0, 50.0)
Version 3: using batches with random shuffling (stochastic gradient descent)#
notice we tune the number of epochs as the number of weights updates increases
#optimization loop
batch_size = 16
epochs = 400
# initialize weights
w.assign(np.random.normal(size=(X.shape[-1],1))*.6)
b.assign(np.random.normal(size=(1,)))
h = []
for epoch in tqdm(range(epochs)):
idxs = np.random.permutation(len(X))
for step in range(len(X)//batch_size+((len(X)%batch_size)!=0)):
X_batch = X[idxs][step*batch_size:(step+1)*batch_size]
y_batch = y[idxs][step*batch_size:(step+1)*batch_size]
gw, gb, loss = get_gradient(w, b, X_batch, y_batch.reshape(-1,1))
w.assign_sub(learning_rate * gw)
b.assign_sub(learning_rate * gb)
h.append([gw.numpy()[0][0], gb.numpy()[0], w.numpy()[0][0], b.numpy()[0], loss.numpy()])
h = np.r_[h]
print (b.numpy(), w.numpy())
100%|███████████████████████████████████████| 400/400 [00:04<00:00, 82.22it/s]
[12.660541] [[-0.7387967]]
predictions = tf.matmul(X,w)+b
rmse = tf.reduce_mean((predictions-y)**2).numpy()
plt.figure(figsize=(15,2));
plt.plot(h[:,-1]); plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse);
plt.ylim(0,50)
(0.0, 50.0)
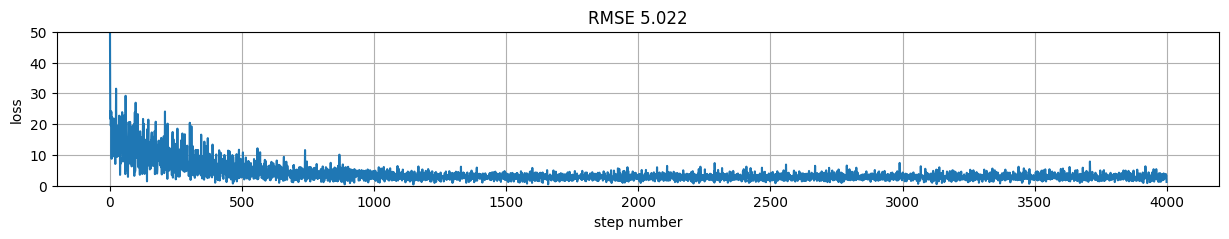
Version 4: packing up with Keras class API and custom SGD#
observe:
the
buildmethod that is called by Keras wheneverinput_shapeis knownwe use
add_weightso that our model weights are known to the Keras model framework (trainable_variables,get_weights, etc.)
see here
class LinearRegressionModel4(tf.keras.Model):
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], 1), initializer='random_normal',
trainable=True, dtype=tf.float32)
self.b = self.add_weight(shape=(1,), initializer='random_normal',
trainable=True, dtype=tf.float32)
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
@tf.function
def get_gradient(self, X, y):
with tf.GradientTape() as t:
loss = tf.reduce_mean( (self(X)-y)**2)
gw, gb = t.gradient(loss, [self.w, self.b])
return gw, gb, loss
def fit(self, X,y, epochs, batch_size=16, learning_rate=0.01):
y = y.reshape(-1,1)
self.h=[]
for epoch in tqdm(range(epochs)):
idxs = np.random.permutation(len(X))
for step in range(len(X)//batch_size+((len(X)%batch_size)!=0)):
X_batch = X[idxs][step*batch_size:(step+1)*batch_size]
y_batch = y[idxs][step*batch_size:(step+1)*batch_size]
gw, gb, loss = self.get_gradient(X_batch,y_batch)
self.w.assign_sub(learning_rate * gw)
self.b.assign_sub(learning_rate * gb)
self.h.append([gw.numpy()[0][0], gb.numpy()[0], w.numpy()[0][0], b.numpy()[0], loss.numpy()])
self.h = np.r_[self.h]
model = LinearRegressionModel4()
observe that we can use the object directly on data to get predictions
model(X[:2])
<tf.Tensor: shape=(2, 1), dtype=float32, numpy=
array([[0.1035429 ],
[0.08904369]], dtype=float32)>
or with the .predict method
model.predict(X[:2])
1/1 [==============================] - 0s 77ms/step
array([[0.1035429 ],
[0.08904369]], dtype=float32)
model.trainable_variables
[<tf.Variable 'linear_regression_model4/Variable:0' shape=(1, 1) dtype=float32, numpy=array([[0.00872076]], dtype=float32)>,
<tf.Variable 'linear_regression_model4/Variable:0' shape=(1,) dtype=float32, numpy=array([0.07025407], dtype=float32)>]
model.get_weights()
[array([[0.00872076]], dtype=float32), array([0.07025407], dtype=float32)]
and fit the model
model.fit(X, y, epochs=400, batch_size=16)
100%|███████████████████████████████████████| 400/400 [00:05<00:00, 76.43it/s]
model.b.numpy(), model.w.numpy()
(array([12.670298], dtype=float32), array([[-0.7236754]], dtype=float32))
predictions = model(X)
rmse = tf.reduce_mean((predictions-y)**2).numpy()
plt.figure(figsize=(15,2));
plt.plot(model.h[:,-1]); plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse);
plt.ylim(0,50)
(0.0, 50.0)
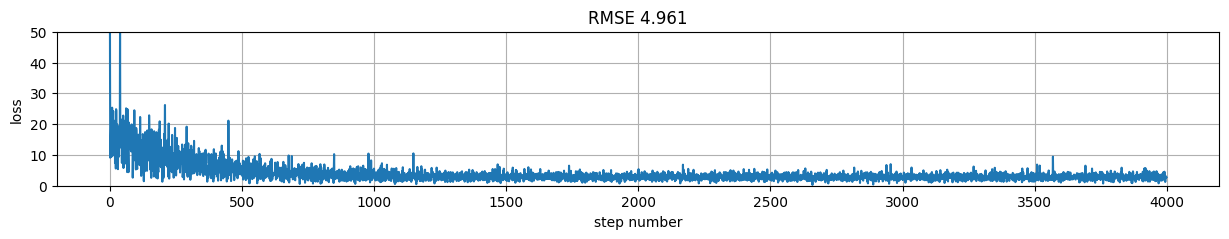
Version 5: Sequential Keras model with standard loop#
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense
def get_model5():
model = Sequential()
model.add(Dense(1, input_shape=(X.shape[-1],), activation="linear"))
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=0.01),
metrics=["mean_absolute_error"],
loss="mse")
# equivalent forms for loss
# loss = lambda y_true, y_pred: tf.reduce_mean((y_true-y_pred)**2))
# loss="mean_squared_error")
# loss=tf.keras.metrics.mean_squared_error)
return model
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(X,y.reshape(-1,1), test_size=0.2)
X_train.shape, X_val.shape, y_train.shape, y_val.shape
((120, 1), (30, 1), (120, 1), (30, 1))
!rm -rf logs
model = get_model5()
tb_callback = tf.keras.callbacks.TensorBoard('./logs', update_freq=1)
model.fit(X_train,y_train, epochs=100, batch_size=5, verbose=0,
callbacks=[tb_callback], validation_data=(X_val, y_val))
model.weights
[<tf.Variable 'dense/kernel:0' shape=(1, 1) dtype=float32, numpy=array([[-0.32406986]], dtype=float32)>,
<tf.Variable 'dense/bias:0' shape=(1,) dtype=float32, numpy=array([11.694233], dtype=float32)>]
history is now logged only per epoch
model.history.history.keys()
dict_keys(['loss', 'mean_absolute_error', 'val_loss', 'val_mean_absolute_error'])
predictions = model(X)
rmse = np.mean((predictions-y)**2)
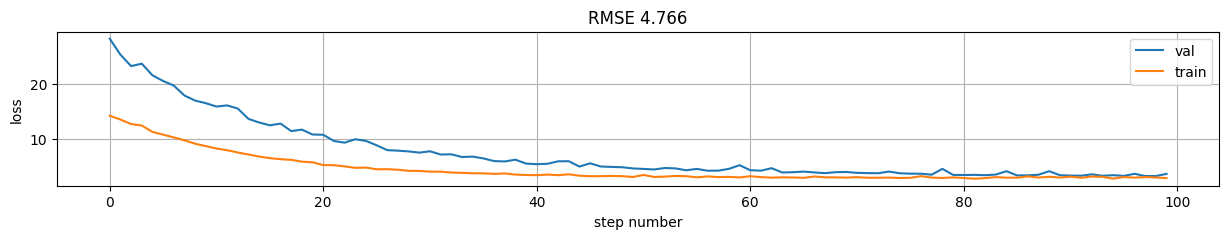
plt.figure(figsize=(15,2));
plt.plot(model.history.history["val_loss"], label="val");
plt.plot(model.history.history["loss"], label="train");
plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse); plt.legend();
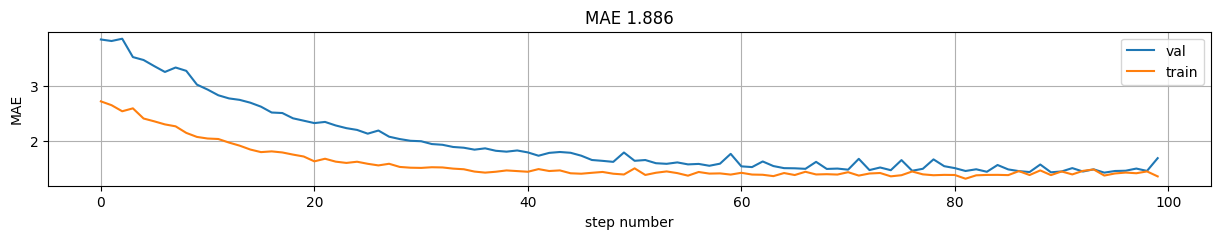
mae = np.mean(np.abs(predictions-y))
plt.figure(figsize=(15,2));
plt.plot(model.history.history["val_mean_absolute_error"], label="val");
plt.plot(model.history.history["mean_absolute_error"], label="train");
plt.xlabel("step number"); plt.ylabel("MAE"); plt.grid();
plt.title("MAE %.3f"%mae); plt.legend();
%tensorboard --logdir logs
Reusing TensorBoard on port 6006 (pid 71551), started 0:22:34 ago. (Use '!kill 71551' to kill it.)
Version 6: Custom model with Keras class API and standard loop#
class LinearRegressionModel6(tf.keras.Model):
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], 1),
initializer='random_normal',
trainable=True, dtype=np.float32)
self.b = self.add_weight(shape=(1,),
initializer='random_normal',
trainable=True, dtype=np.float32)
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
model = LinearRegressionModel6()
!rm -rf logs
model = LinearRegressionModel6()
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=0.02),
loss="mse", metrics=['mean_absolute_error'])
tb_callback = tf.keras.callbacks.TensorBoard('./logs', update_freq=1)
model.fit(X_train,y_train, epochs=100, batch_size=5, callbacks=[tb_callback],
verbose=0, validation_data=(X_val, y_val))
<keras.src.callbacks.History at 0x7fce31557c40>
You can also define your own optimizer (see this blog)
model.b.numpy(), model.w.numpy()[0]
(array([11.829514], dtype=float32), array([-0.68691593], dtype=float32))
predictions = model(X)
rmse = np.mean((predictions-y)**2)
plt.figure(figsize=(15,2));
plt.plot(model.history.history["loss"], label="train");
plt.plot(model.history.history["val_loss"], label="val");
plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse); plt.legend();
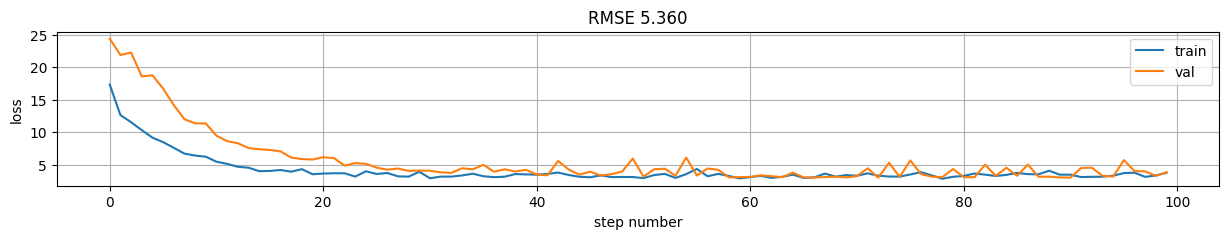
Version 7: Using train_step \(\rightarrow\) control loss and gradients on a custom model.#
class LinearRegressionModel7(tf.keras.Model):
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], 1),
initializer='random_normal',
trainable=True, dtype=np.float32)
self.b = self.add_weight(shape=(1,),
initializer='random_normal',
trainable=True, dtype=np.float32)
self.loss_fn = tf.keras.metrics.MeanSquaredError()
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
def test_step(self, data):
# here we implement loss by hand
return {'loss': tf.reduce_mean((self(X)-y)**2) }
@tf.function
def train_step(self, data):
X,y = data
loss_fn = lambda y_true, y_preds: tf.reduce_mean((y_true-y_preds)**2)
with tf.GradientTape() as tape:
# we use tf.keras loss function (equivalent to test_step)
loss_fn = tf.keras.metrics.mean_squared_error
loss = tf.reduce_mean(loss_fn(y, self(X)))
grads = tape.gradient(loss, self.trainable_variables)
self.optimizer.apply_gradients(zip(grads, self.trainable_variables))
return {'loss': loss}
model = LinearRegressionModel7()
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=0.02))
#model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=0.035))
model.fit(X_train,y_train, epochs=400, batch_size=5, verbose=0, validation_data=(X_val, y_val))
<keras.src.callbacks.History at 0x7fce23215fd0>
[i.numpy() for i in model.trainable_variables]
[array([[-0.49167478]], dtype=float32), array([11.938187], dtype=float32)]
predictions = model(X)
rmse = np.mean((predictions-y)**2)
plt.figure(figsize=(15,2));
plt.plot(model.history.history["loss"], label="train");
plt.plot(model.history.history["val_loss"], label="val");
plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse); plt.legend();
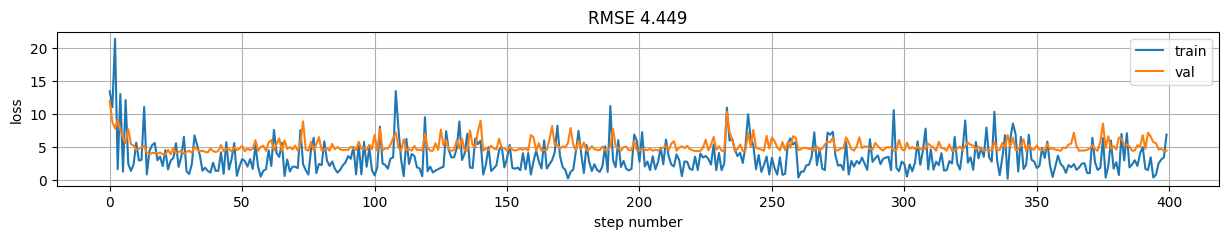
Version 8: Using train_step \(\rightarrow\) control loss and gradients on a standard model.#
observe that:
we use a standard
Denselayer,we use a custom loss function and
optimizer.apply_gradients
class CustomModel(tf.keras.Model):
def test_step(self, data):
return {'loss': tf.reduce_mean((self(X)-y)**2)}
@tf.function
def train_step(self, data):
X,y = data
with tf.GradientTape() as tape:
y_pred = self(X, training=True)
loss_value = tf.reduce_mean((y_pred-y)**2)
grads = tape.gradient(loss_value, self.trainable_variables)
self.optimizer.apply_gradients(zip(grads, self.trainable_variables))
return {'loss': loss_value}
from tensorflow.keras.layers import Dense, Input
def get_model8():
inputs = tf.keras.layers.Input(shape=(1))
outputs = tf.keras.layers.Dense(1, activation="linear")(inputs)
model = CustomModel(inputs, outputs)
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=0.02))
return model
our custom loop (for any model !!!)
model = get_model8()
model.summary()
Model: "custom_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 1)] 0
dense_1 (Dense) (None, 1) 2
=================================================================
Total params: 2 (8.00 Byte)
Trainable params: 2 (8.00 Byte)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
model.weights
[<tf.Variable 'dense_1/kernel:0' shape=(1, 1) dtype=float32, numpy=array([[1.5373226]], dtype=float32)>,
<tf.Variable 'dense_1/bias:0' shape=(1,) dtype=float32, numpy=array([0.], dtype=float32)>]
model.trainable_variables
[<tf.Variable 'dense_1/kernel:0' shape=(1, 1) dtype=float32, numpy=array([[1.5373226]], dtype=float32)>,
<tf.Variable 'dense_1/bias:0' shape=(1,) dtype=float32, numpy=array([0.], dtype=float32)>]
model.fit(X_train,y_train.reshape(-1,1), epochs=400, batch_size=5, verbose=0, validation_data=(X_val, y_val))
<keras.src.callbacks.History at 0x7fce315a8ac0>
model.trainable_variables
[<tf.Variable 'dense_1/kernel:0' shape=(1, 1) dtype=float32, numpy=array([[-0.5964833]], dtype=float32)>,
<tf.Variable 'dense_1/bias:0' shape=(1,) dtype=float32, numpy=array([11.866811], dtype=float32)>]
predictions = model(X)
rmse = np.mean((predictions-y)**2)
plt.figure(figsize=(15,2));
plt.plot(model.history.history["loss"], label="train");
plt.plot(model.history.history["val_loss"], label="val");
plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse); plt.legend();
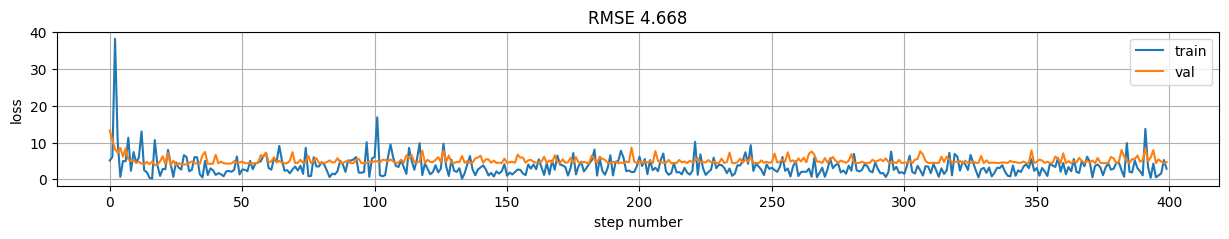
Version 9: using train_on_batch \(\rightarrow\) control data#
epochs = 400
batch_size = 5
model = get_model8()
h = []
for epoch in tqdm(range(epochs)):
idxs = np.random.permutation(len(X))
for step in range(len(X)//batch_size+((len(X)%batch_size)!=0)):
X_batch = X[idxs][step*batch_size:(step+1)*batch_size]
y_batch = y[idxs][step*batch_size:(step+1)*batch_size]
model.train_on_batch(X_batch, y_batch)
h.append(model.test_step([X_batch, y_batch])['loss'])
100%|███████████████████████████████████████| 400/400 [00:48<00:00, 8.28it/s]
model.trainable_variables
[<tf.Variable 'dense_2/kernel:0' shape=(1, 1) dtype=float32, numpy=array([[-0.17166868]], dtype=float32)>,
<tf.Variable 'dense_2/bias:0' shape=(1,) dtype=float32, numpy=array([10.048985], dtype=float32)>]
predictions = model(X)
rmse = tf.reduce_mean((predictions-y)**2).numpy()
plt.figure(figsize=(15,2));
plt.plot(h); plt.xlabel("step number"); plt.ylabel("loss"); plt.grid();
plt.title("RMSE %.3f"%rmse);
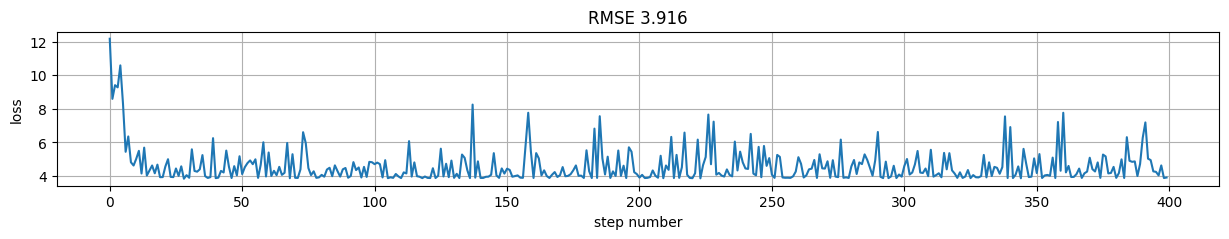
Note that we can still have access to gradients with respect to the any of the layers in the Computational Graph (if connected)#
with tf.GradientTape() as tape:
y_pred = model(X, training=True)
loss_value = tf.reduce_mean((y_pred-y)**2)
grads = (tape.gradient(loss_value, model.layers[1].weights))
print(grads)
[<tf.Tensor: shape=(1, 1), dtype=float32, numpy=array([[-1.664384]], dtype=float32)>, <tf.Tensor: shape=(1,), dtype=float32, numpy=array([-0.20064919], dtype=float32)>]